Abdulaev N.G.
|
Professor, Doctor of Science Орден Дружбы Народов, Медаль «За трудовую доблесть», Государственная премия в области науки и техники, Премии АН СССР и РАН имени М.М. Шемякина |
Nazhmutin Gadzhymagomedovich Abdulaev (1941-2018) completed his studies at the Moscow Lomonosov Institute of Fine Chemical Technology, and, from 1967 to 1994, worked at the Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry. From 1994, N. G. Abdulaev worked in a professorial capacity at the University of Maryland in Rockville, Maryland, United States of America.
List of trainees and PhD students
Artamonov ID, Bogachuk AS, Gaidarov IO, Grozeva LM, Dergachev AE, Eganyan E, Kakuev DL, Karaschuk GN, Kiselev AV, Klezovich ON, Kolosov VL,Kutuzov MA, Lobanov NA, Popov VI, Shmukler BE, Solovieva OV, Suslov ON, Taneva SG, Vasileva CV, Zargarov AA, Zolotarev AS, Yakhyaev AV.
- Alexander von Humbolt award (Germany, 1992)
- State premium of USSR and Russian Federation in the field of science and technology. Received for a cycle of works towards exploration of transmembrane ionic channels (1986)
- Recipient of the Order of International Friendship (1986)
- Premium of the Academy of Sciences of the USSR and Russian Academy of Sciences. Received for publication of work titled “Bioorganic Chemistry of Rhodopsins.” (1983)
- Medal “For Labour Valour” (1981)
Education
| Period | Coyntry, city | University | Additional info |
|---|---|---|---|
| 1970–1985 | Russia, Moscow | Shemyakin Institute of Bioorganic Chemistry | Doctor of Chemical Sciences |
| 1970–1973 | Russia, Moscow | Shemyakin Institute of Bioorganic Chemistry | Candidate of Chemical Sciences (Ph.D.) |
| 1962–1967 | Russia, Moscow | Lomonosov Institute of Fine Chemical Technology | Master's degree in Chemical Engineering (Diploma) |
| 1956–1960 | Russia, Mahachkala | Medical College | Diploma in nursing |
IBCh positions
| Head | Scientific units |
Titles
| Professor | |
| Doctor of Science (Chemistry) | |
Awards
| 1986 | Орден Дружбы Народов | |
| 1981 | Медаль «За трудовую доблесть» | |
| 1986 | Государственная премия в области науки и техники | |
| 1983 | Премии АН СССР и РАН имени М.М. Шемякина |
Scientific interests
Prof. N. G. Abdulaev’s research was primarily focused on investigation of the structure-function relation in proteins involved in signal transduction and cGMP pathways in retina. Dr. Abdulaev’s work particularly emphasized the study of rhodopsin, a member of the superfamily of G-protein coupled receptors (GPCR’s). His scientific findings laid a strong foundation in understanding the principles of rhodopsin assembly and folding. N. G. Abdulaev made an invaluable contribution towards the development of a methodology designed for the analysis of the primary structure and topography of integral membrane proteins. This methodology is still widely used.
Scientific results
Structure-Functional Studies of Aspartate Aminotransferase
Structure-Fuctional studies of Bacteriorhodopsin
Under the supervision of Yu. A. Ovchinnikov, N. G. Abdulaev determined the sequence of the integral membrane protein bacteriorhodopsin of the so-called purple membranes of some salt-loving bacteria – Halobacteria. Bacteriorhodopsin is a chromoprotein and has a retinal residue in all-trans or 13-cis configuration as a light-sensitive system. In Halobacteria, bacteriorhodopsin functions as a light-driven proton pump generating electrochemical gradient of protons that is subsequently converted into chemical energy.
One of the first obstacles in structural analysis of integral membrane proteins is their near-complete insolubility in water-based buffer systems. Since this essentially excluded the application of proteolytic enzymes in their traditional ways, the use was made of splitting the polypeptide chain mostly by chemical methods. The resulting fragments were isolated by gel permeation chromatography either in concentrated formic acid or 8 M guanidinium chloride solution.
During the course of this work, a number of highly important findings were made as to the topography of the molecule in the membrane. A thorough analysis of the distribution of hydrophobic and hydrophilic amino acid residues revealed the high content of ‘hydrophobic’ amino acids (-66% of the total number) and their ‘clustering’ in definite regions of the polypeptide chain. The hydrophobic clusters represent seven transmembrane segments, while hydrophilic residues form short sequences connecting the segments.
Limited proteolysis also yielded valuable knowledge of the topological organization of bacteriorhodopsin. Enzymatic treatment of bacteriorhodopsin in purple membranes and inside-out vesicles lead to splitting only in the regions exposed to the exterior whereas the inaccessible regions of the protein deeply embedded in the membrane withstood attack. The findings indicated that the N- terminus of bacteriorhodopsin is localized on the surface of the membrane while its C-terminus is situated in the cell cytoplasm. Treatment with enzymes of broad specificity, particularly papain, revealed four areas of polypeptide chain exposed on both surfaces of the cell membrane. It was also found that several parts of the molecule are not essential for proton pumping activity. Furthermore, the protein possessed an ability to refold after complete denaturation. The full proton- pumping activity has been restored after complete delipidation and following reconstitution of bacteriorhodopsin into liposomes.
Immunochemical techniques represented a powerful tool in refining the spatial organization of bacteriorhodopsin in the membrane. Monoclonal antibodies directed against various peptide fragments of the protein were used for identification of the exposed regions. It was shown that 3 regions in bacteriorhodopsin molecule are exposed on the cell surface while 4 others are intramembranous.
Determination of the retinal location in bacteriorhodopsin was also of utmost importance. The study of bacteriorhodopsin and its derivatives treated with NaBH4 resulted in conclusion that the retinal residue is bound to the C-terminal part of the protein at Lys (216).
Taking together, the studies, begun in 1976, resulted in 1978 in the complete structural elucidation of bacteriorhodopsin, the first of the true membrane proteins for which this has been successfully accomplished. The first model for the polypeptide chain arrangement of bacteriorhodopsin in the membrane was proposed. The results have opened wide perspectives for the studies of other membrane proteins.
Structure-Fuctional Studies of Bovine and Octopus Rhodopsin
In the late 1970s, N. G. Abdulaev focused on studies of rhodopsin, the light-sensitive pigment from photoreceptor cells of vertebrate retina. Like bacteriorhodopsin, rhodopsin is an integral membrane protein. And likewise, it contains a retinal moiety as a light-sensitive component. Thus, it is classified as a chromoprotein. Rhodopsin plays a key role in transmitting the visual signal along the optical nerve to the visual cortex in the brain.
In structural analysis of bovine rhodopsin, N. G. Abdulaev relied on the strategy and tactics used earlier for bacteriorhodopsin. The complete sequence of rhodopsin was determined in 1982. N. G. Abdulaev developed a highly specific procedure for limited proteolysis of the protein in native and apo-membranes. Several monoclonal antibodies directed against rhodopsin were generated to investigate its topology and mechanism of activation.
Treatment of rhodopsin in photoreceptor disc membranes of native and inverted orientation with proteolytic enzymes refined the length and location of the parts polypeptide chain exposed to intradiscal space and the outer membrane surface. The results also pointed to the peptide loops essential for providing an interface between rhodopsin and downstream members of the signaling pathway.
Further studies with monoclonal antibodies generated against activated state of rhodopsin, demonstrated that transition to activated Meta II state exposes cytoplasmic segment of transmembrane helix VII in rhodopsin.
Among the major findings of this work were the following: (i) Polypeptide chain of rhodopsin was shown to consists of 348 amino acid residues; (ii) Retinyl residue is bound to Lys (296); (iii) two oligosaccharide chains are attached to Asn(2) and Asn(15); (iiii) Rhodopsin has 2 palmitoyl groups covalently-bound via thioester bound to Cys(322) and Cys(323). Removal of these fatty acids by hydroxylamine or other reducing agents does not affect ability of rhodopsin to form active molecule or activate G-protein.
The data resulted in proposal of a model for rhodopsin topography (Fig. 1). Its principal feature is the presence of 7 membrane-spanning rods interconnected by intermittent hydrophilic fragments. The roads contain - 60% of all the amino acid residues and are essentially in a-helical conformation.
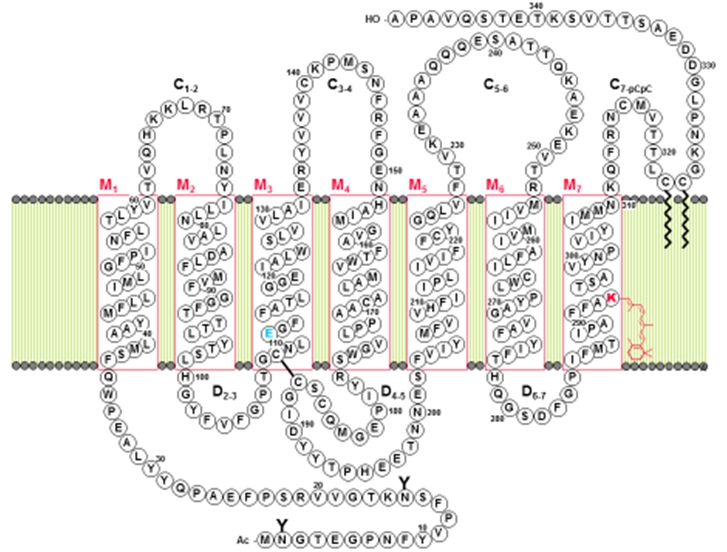
Fig.1 Disposition of the rhodopsin polypeptide chain in photoreceptor membrane
Despite similarities in the topographies of animal and bacterial rhodopsins, there is no convincing evidence in favor of an evolutionary interrelation between them. Computer analysis of sequences of both rhodopsins showed that these proteins have no identical sequences longer than 5 amino acid residues.
In the late 1990th, N. G. Abdulaev was aimed with elucidation of the primary structure of rhodopsin from the octopus Paroctopus depeini. The sequence was determined by parallel analysis of the protein and corresponding cDNA. It contains 455 residues of molecular mass 50,234 Da. Octopus rhodopsin has Lys-306 as the site of covalent attachment of 11-cis retinal. The polypeptide chain is divided into several alternating hydrophilic and hydrophobic sequences, the latter being much more extended than those in bovine rhodopsin. A characteristic feature of polypeptide chain is its long carboxyl-terminal tail containing large number of Pro- and Gln- amino acid residues. There are two potential glycosylation cites closer to the N-terminus of the protein. The Cys-337 and Cys-338 are also conserved in the C-terminal tail suggesting their involvement in posttranslational fatty acid acylation. It also contains several threonine and serine residues and thus meets the requirement of being phosphorylated in a light-dependent manner. The data resulted in proposal of the model of topological organization of octopus rhodopsin in the membrane (Fig. 2). It depicts seven transmembrane regions connected by polypeptide loops on both sides of the membrane.
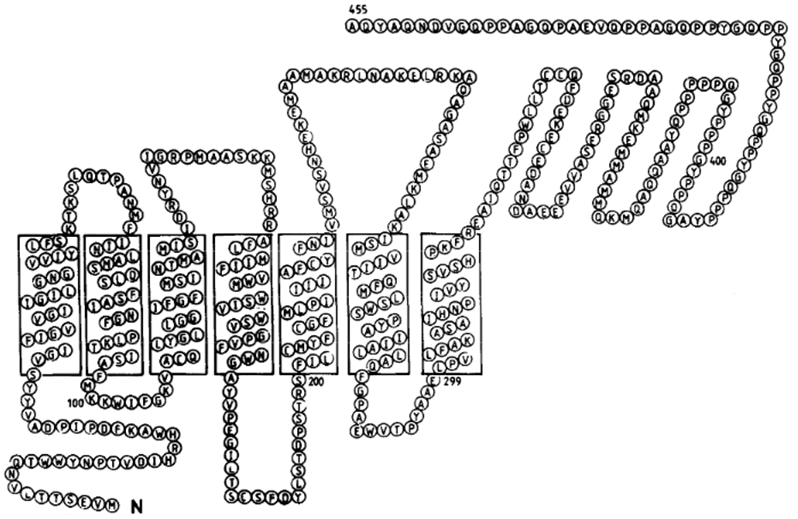
Fig.2 The transmembrane topology of octopus rhodopsin
After Yu. A. Ovchinnikov’s passing, N. G. Abdulaev continued study on rhodopsin in collaboration with Kevin Ridge. The subject of their study was to clarify details of rhodopsin folding, assembly and function by means of using the protein fragments. Several complementary bovine opsin fragments were examined whether they contain sufficient information to independently fold, insert into a membrane, and assemble into a functional pigment. The studies identified an amino-terminal fragment (1-232), which folds and inserts into a membrane only after coexpression with its complementary carboxyl-terminal fragment (233-348). To further characterize this interaction, the fragment (1-232) production was examined upon coexpression with carboxyl-terminal fragments of varying length and / or amino acid composition. It was shown that although the fragment (1-232) production was independent of the third cytoplasmic loop (amino acids 233-249) and carboxyl-terminal tail (amino acids 313-348), both the sixth and the seventh transmembrane helices were essential. Mutations of the highly conservative Pro-267 residue presumably associated with retinitis pigmentosa showed that in some cases, the presence of the third cytoplasmic loop within the sixth fragment is detrimental to the fragment (1-232) expression. It was suggested that conformation of the third cytoplasmic loop, if present, is important for establishing the proper contacts required for protein folding, membrane insertion, assembly and/or function. In further studies on rhodopsin, an attempt was made to reconstruct the interacting surface(s) for the proteins involved in visual transduction and desensitization. For that, fragments corresponding to the cytoplasmic loops and/or the carboxyl-terminal tail of bovine opsin either singly, or in combination, were inserted onto a surface loop in thioredoxin scaffold. The results showed that the construct with the second and third cytoplasmic loops independent on the chromophore and the connecting transmembrane helices effectively activated G-protein.
Expression of Rhodopsin in Yeast
One of the constraints in structure-functional analysis of rhodopsin exists, in part, because of lack of effective expression system capable of producing large amounts of functional protein. N. G. Abdulaev was the first to demonstrate applicability of the methylotrophic yeast P. pastoris for this purpose. The bovine opsin gene was placed downstream from the P. pastoris alcohol oxidase gene promoter and fused at its N-terminus to the acid phosphatase secretion signal. This resulted in one copy of rhodopsin gene to be integrated into the yeast genome. The expression level of some transformants corresponded to 0.3 mg of opsin per liter of the cell culture. The expressed protein, similar to the native rhodopsin, had high mannose N-glycosylation, MW of 37KDa and was able to interact with 11-cis retinal to form the rhodopsin chromophore. The metharhodopsin II-mediated G-protein activating potential of the expressed rhodopsin was also similar to that of the native rhodopsin.

















