Laboratory of structural biology of ion channels
In the picture from left to right: Mikhail Yu. Myshkin, Zakhar O. Shenkarev, Andrey V. Tsarev, Milita V. Kocharovskaya, Alexander S. Paramonov.
Our laboratory studies membrane receptors and ion channels, and also compounds acting upon them (toxins, endogenous ligands). For studies of structure and molecular mechanisms of action we apply modern methods of structural biology: NMR-spectroscopy and cryo-electron microscopy. At the moment we are studying mechanisms of interaction between toxins and voltage-gated K+ and Na+ channels (Kv and Nav), and also the structures of protein ligands of nicotinic acetylcholine receptor (nAChR). Structural studies of membrane receptors and channels are necessary for development of new methods of diagnostics and therapy of deceases of nervous, cardiovascular and muscular systems. Besides, we study structures and mechanisms of action of antibiotics and defensive peptides of plants and animals. These studies are important for development of new drugs for treatment of infectious deceases.

Fig 1. Structure of complex of human muscle Nav1.4 channel and Hm-3 toxin from Heriaeus melloteei spider venom. Structure was calculated based on NMR data. (A) Complex of Hm-3 (blue/purple) and first voltage-sensing domain (DI) of Nav1.4 channel (pale-yellow/red). Side view from lipid bilayer. (В) Toxin-channel complex. View from outside cell on the membrane surface.
Our laboratory carried out NMR studies of the structure and dynamics of voltage-sensing domains of several K+ and Na+ channels, including Nav1.4 channel from human muscles. Dysfunctions of Nav1.4 channel cause disorders of musculoskeletal system, such as paralysis, myasthenic syndrome and myotonia. For the first time, “unusual” fluctuations in the structure of domains occurring with characteristic times in the μs-ms range, possibly the prototype of the structural rearrangements occurring during voltage-dependent activation, were characterized. It is shown that the domains of Na+ channels have a greater conformational plasticity compared to the domains of K+ channels. This property probably leads to a faster response of the Nav channels when the transmembrane potential changes. We have studied VSTx1 and Hm-3 toxins from spider venom, which by binding to the membrane patch surrounding the ion channel block voltage-dependent activation. Based on the experimental data of NMR spectroscopy, together with the Laboratory of Molecular Instruments for Neurobiology IBCh RAS a model of the toxin complex Hm-3 with Nav1.4 channel of human skeletal muscles was constructed. The obtained structural data opens up the possibility of further pharmacological developments.
Currently, together with the Biological Department of Moscow State University, using electron microscopy methods, we are investigating the spatial structure of full-sized Kv7.1 channel of human cardiac muscle. Dysfunctions of this channel lead to the development of hereditary arrhythmias – a syndrome of an extended QT interval.
Together with the group of Bioengineering of Neuromodulators and Neuroreceptors of IBCh RAS, using the methods of NMR spectroscopy, we determined the structures of several “three-loop” proteins acting on the ion channel of the nicotinic acetylcholine receptor (nAChR). The spatial structures of the WTX toxin from cobra venom, Lynx1 and Lypd6 regulatory proteins from the human nervous system, and SLURP-1 and SLURP-2 proteins produced by human epithelium cells were determined. These molecules have prospects for the development of new drugs aimed at the treatment of neurodegenerative diseases.
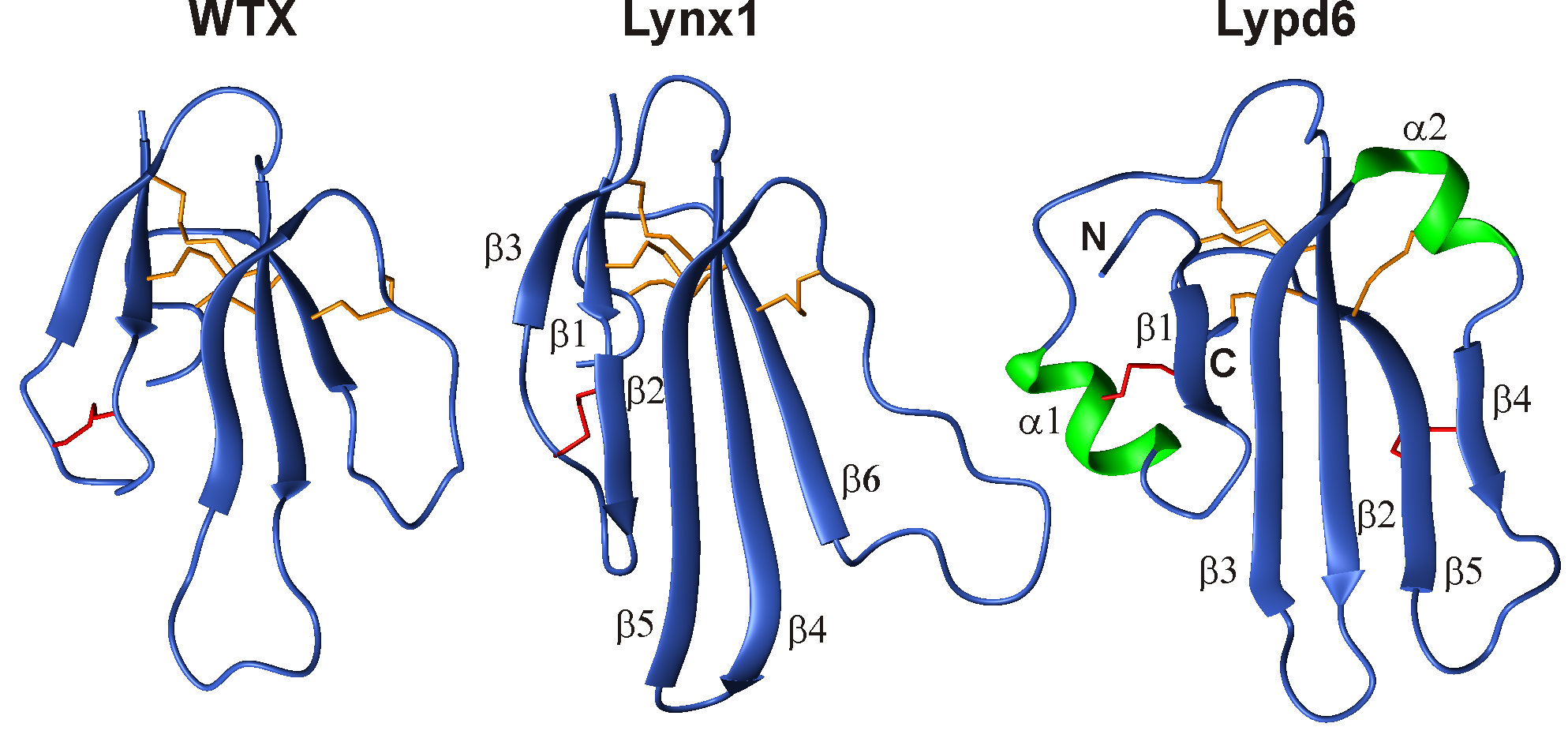
Fig 2. Spatial NMR structures of three-loop proteins, acting upon nicotinic acetylcholine receptors. WTX – toxin from venom of Naja kaouthia cobra. Lynx1 and Lypd6 – regulating proteins of human nervous system.
Using the methods of NMR spectroscopy, our laboratory together with the Educational and Scientific Center of IBCh RAS examines the structure and mechanism of action of new antibiotics and defensive peptides of plants and animals. Most of the studied molecules exhibit increased affinity for the membranes of bacterial cells and are able to form pores and ion channels, which lead to the death of the target cell. We have determined the spatial structure and dynamics of the following molecules: two-component lantibiotic lichenicidin, antiamoebin antibiotic, an antimicrobial peptide arenicine from marine worm, antimicrobial peptide aurelin from jellyfish, defensin peptide from lentils, a lipid-transporting protein from lentils in complex with lipids.
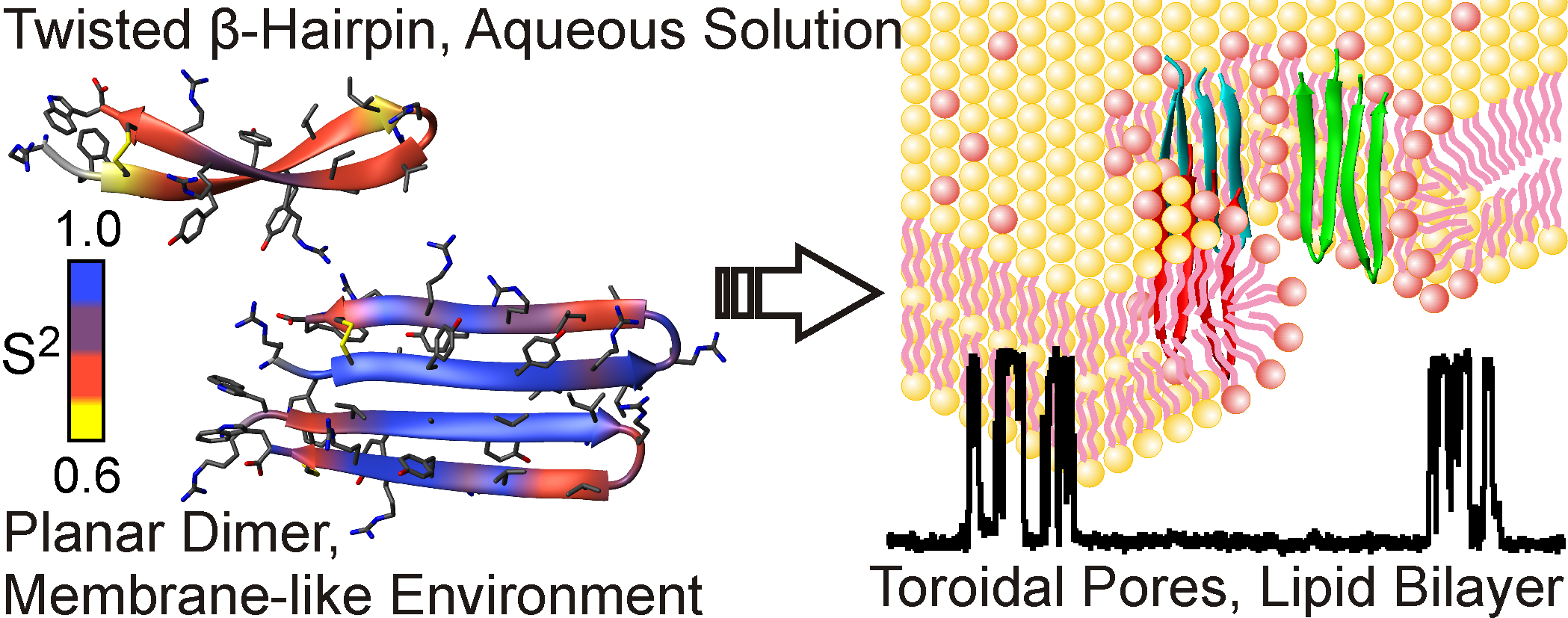
Fig 3. Spatial structure and conformational plasticity of antimicrobial peptide arenicin monomer in water and dimer in membrane environment. Formation of toroidal pores in bacterial membranes by arenicin is shown on the right.
The laboratory is also developing new approaches for studying membrane biomolecules. One of the promising areas is the use of nanoparticles based on high-density lipoproteins (nanodisc). We first demonstrated the possibility of using nanodiscs as a medium for NMR studies of membrane proteins and membrane-active peptides, investigated the possibility of using nanodisks in cell-free synthesis systems to obtain functionally active forms of membrane proteins, as well as the possibility of using nanodiscs for folding membrane proteins in vitro.

Fig 4. The addition of preformed nanodiscs to the translational mixture of the cell-free synthesis system makes it possible to obtain functionally active membrane proteins available for studies by NMR spectroscopy.
Our laboratory is developing new approaches to establish the chemical structure and study the mechanisms of transformation into nitrogen-rich heterocyclic compounds. One of the developed approaches is based on measuring and analyzing “small” (amplitude from 1 to 0.01 Hz) 13C-15N and 1H-15N spin-spin coupling constants in compounds selectively labeled with stable 15N isotope. The application of this approach allowed us to study the processes of azido-tetrazole tautomerism in azido-triazines and azido-pyrimidines.

Fig 5. Analysis of «small» 13С-15N spin-spin coupling constants in compounds labeled with stable 15N isotope, allows to determine the structure of terazole form in azido-triazines and azido-pyrimidines.
Our laboratory works in collaboration with subdivisions of IBCh RAS:
- Group of bioengineering of neuromodulators and neuroreceptors (Head – Ekaterina N. Lyukmanova)
- Laboratory of biomolecular NMR-spectroscopy (Head – Alexander S. Arseniev)
- Laboratory of Molecular Instruments for Neurobiology (Head – Alexander A. Vassilevski)
- Educational and Scientific Center IBCh RAS (Head – Tatiana V. Ovchinnikova)
- Laboratory of molecular design and synthesis IBCh RAS (Head – Vladimir A. Korshun)
Our laboratory collaborates with Russian scientific and educational organizations:
- Structural Biotechnology Group Bioengineering department at Lomonosov Moscow State University (Head – Olga S. Sokolova)
- Laboratory of Magnetic Resonance, Institute «International Tomography Center» SD RAS, Novosibirsk
- Department of Organic and Biomolecular Chemistry, Ural Federal University
Foreign collaborations:
- Sobolevsky Lab, Department of Biochemistry and Molecular Biophysics, Columbia University (USA) (Head - Alexander Sobolevsky)
The group of structural biology of ion channels was founded in 2015 as a part of the program of Presidium RAS “Molecular and Cell Biology” (de facto the group existed since 2008). In 2019 the group was given laboratory status.
- Structural studies of human K+ and Na+ channels and mechanisms of action of toxins, affecting voltage-dependent activation.
- Studies of interconnection between structure and function of "three-loop" proteins – ligands of nicotinic acetylcholine receptor (nAChR).
- Studies of sctructure and mechanisms of action of new antibiotics and defensive antimicrobial peptides.
- Development of new membrane-mimicking media on the basis of high-density lipoproteins (nanodiscs) for production and ctructural and functional studies of membrane proteins and peptides.
- Development of new NMR techniques for determination of structure and mechanisms of transformation of nitrogen-rich heterocyclic compounds.
| Fullname | Position | Contacts |
|---|---|---|
| Zakhar Shenkarev, Prof.RAS, D.Sc | pr. r. f. | zakhar-shenkarev@yandex.ru |
| Leichenko E.V. | s. r. f. | |
| Alexander Paramonov, Ph.D. | s. r. f. | |
| Shabel'nikov S.V. | s. r. f. | |
| Kvetkina A.N. | j. r. f. | |
| Maxim Zaigraev | j. r. f. | maximzaigraev@gmail.com |
| Elizaveta Chernaya | res. eng. | |
| Milita Kocharovskaya | j. r. f. | |
| Kovalenko E.A. | res. eng. | |
| Pavel Mironov | j. r. f. | |
| Stanislav Gorelov | res. eng., PhD stud. | |
| Stanislav Gorelov | res. eng., PhD stud. | |
| Ivannikov A.D. | t. q. - lab. as. | |
| Sergey Oreshkov | t. q. - lab. as. | |
Previously worked here | ||
| Karlova M.G. | ||
| Marilovceva E.V. | ||
| Mikhail Myshkin | ||
| Popova A.S., Ph.D. | ||
| Tsarev A.V. | ||
| Zanyatkin I.A., Ph.D. | ||
| Chernikov A.M. | ||
| Horev A.S. | ||
| Loktyushov E.V. | ||
| Surkova E.A. | ||
| Litvinov D.S. | ||
| Reznikova O. | ||
| Ulanova P.V. | ||
 Loading...
Loading...Scientific projects
 Loading...
Loading... Loading...
Loading...
