Laboratory of X-ray study
The Laboratory is involved in study of three dimensional (3D) structure of the protein-peptide nature compounds by high resolution X-ray methods supported by molecular mechanics/dynamics/graphics and bioinformatics methods. The Laboratory studies proteins of different functional nature with emphasis on the structure-functional aspects of specificity of the ligand/substrate recognition and binding.
The Laboratory collaborates with the Macromolecular Crystallography Laboratory of the National Cancer Institute of USA (Argonne, IL, USA). The laboratory of X-ray study was organized in 1990 from the X-ray group functioning since 1972.

Laboratory employees.
In the present time, much attention in our lab is paid to structural study of the fluorescent proteins (FP) used as molecular bionanomarkers in cell biology, biotechnology and biomedicine for visualization and monitoring the internal processes within cells or whole organisms. A big series of FPs, emitting in green, yellow, red and far red spectral regions, has been studied in our Lab by high resolution X-ray methods. The following analysis of the structure-functional relation allowed to explain many experimentally observed properties and to design new mutant fluorescent variants with improved photophysical characteristics. The obtained results of the Laboratory are expanding considerably the structural base for rational design of the advanced fluorescent biomarkers for practical application.
The 3D structures of the cyclic depsipeptide ionophores differing by cycle size, nature and configuration of residues helped to understand the important details of the metal ion binding specificity and mechanism of the ion transport through biological membranes.
In collaboration with Hauptman-Woodward Medical Research Institute (Buffalo, USA) a number of research projects has been performed under the title “Rational proteomics of short chain dehydrogenases”. A general approach for identification of the 3D pattern of residues (fingerprints) responsible for the protein fold, cofactor and substrate binding was developed for this family.
A big series of FPs, emitting in green, yellow, red and far red spectral regions, has been studied in our lab by high resolution X-ray methods. The following analysis of the structure-functional relation allowed to explain many experimentally observed properties and to design new mutant fluorescent variants with improved photophysical characteristics.
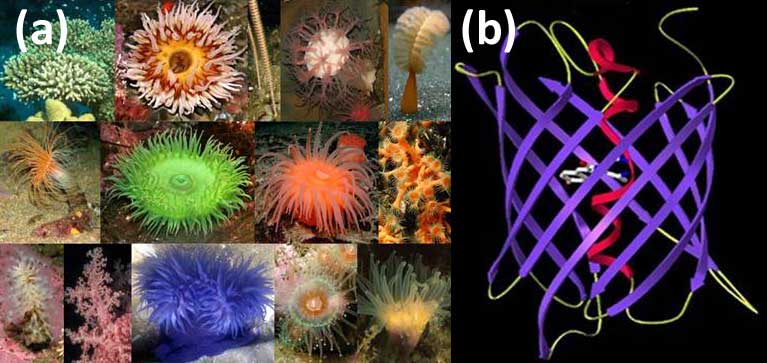
Selected species of the marine button polyps — the source of fluorescent proteins (FP) (a); The principal structural fold of FPs is an 11-stranded β-barrel and a chromophore (matured from the three residue sequence) embedded in the middle of an internal α-helix going along the β-barrel axis (b).

Crystals of the fluorescent proteins for X-ray study
Unic examples of fluorescent proteins obtained in the laboratory
I. Study of the far red monomeric FP mKate has showed that the observed pH dependence of fluorescence is a consequence of cis-trans isomerization of the internal chromophore. Based on its 3D structure the new gene engineered variant mKate_S158A with almost 2 fold brighter fluorescence has been rationally designed. Currently, the photophysical characteristics of this variant succeed significantly over those of other known monomeric fluorescent biomarkers.
Structural study of the highly toxic red and orange FPs, KillerRed and KillerOrange, allowed to determine the chromophore adjacent key amino-acid residues participating in generation of the active oxygen forms, responsible for the phototoxic effect.

Amino acid environment of the internal chromophore in highly toxic red fluorescent protein KillerRed. Hydrogen bonds (≤3.3 Å) are shown as blue dashed lines, waters (W) as red spheres and van der Waals contacts (≤3.9 Å) as black “eyelashes”.
II. The intermediate form of chromophore biosynthesis has been observed in crystal structure of the colorless nonfluorescent FP, aceGFP-G222E. This new experimentally observed structure of the immature chromophore, characterized by the non-coplanar arrangement of the imidazolinone and phenolic rings, where cyclization of the protein backbone has occurred, but Tyr66 chromophore still stays in the native, non-oxidized form, with Cα and Cβ atoms in sp3 hybridization.

The trapped intermediate state (form III) for GFP chromophore biosynthesis found in the crystal structure of aceGFP-G222E
III.

Red Fluorescent Protein laRFP from a lancelet. Chromophore Gly58-Tyr59-Gly60 (GYG; shown in yellow) in 2Fo-Fc electron density (density cutoff ρ= 2.0σ). laRFP showing the presence of the covalent bond between chromophore (Tyr59)Cβ and proximal (Tyr62)O. Potential proton acceptor Asp142 forms H-bond (shown in dashed red line), mediated by water molecule (W), with the Tyr62 hydroxyl. Arg89 forms H-bond with the Tyr62 hydroxyl that assumingly may facilitate proton transfer to Asp142 prior to reaction.
IV.

Канал простирающейся вдоль β-бочонка заполнен цепочкой из связанных водородными связями молекул воды, выполняющей роль транспортной системы для фотогенерируемых токсичных супероксид анионов.
V. For the first time, the three-dimensional structure of the native green and UV irradiated photoconverted red forms of DendFP (Dendronephthya sp.) has been determined, the latter showing cleavage of the main chain before chromophore

Chromophore structure (His62-Trp63-Gly64) in the green form of DendGFP (a) and the red form of DendRFP (b).
VI. In international collaboration three new bright far-red and near infrared genetically engineered biomarkers (from plant photoreceptors - phytochromes), providing high permeability of emission through biological tissues - miRFP670 (em ~ 670nm ), miRFP703 (703 nm) and miRFP709 (709 nm), have been studied by X-ray method with resolution 1.33, 1.35 and 1.34Å, respectively. Three-dimensional structure and structure-functional relations of these biomarkers have been established

Superimposed structures of miRFP703 (in green) and miRFP709 (in red) showing chromophores in the binding pocket
| Fullname | Position | Contacts |
|---|---|---|
Previously worked here | ||
| Vladimir Pletnev, D.Sc | ||
| Tsygannik I.N., Ph.D. | ||
| Igor' Artem'ev | ||
| Ekaterina Goryacheva, Ph.D. | ||
| Svetlana Arhipova | ||
| Rossohin A.V. | ||
 Loading...
Loading...Scientific projects
 Loading...
Loading... Loading...
Loading...
